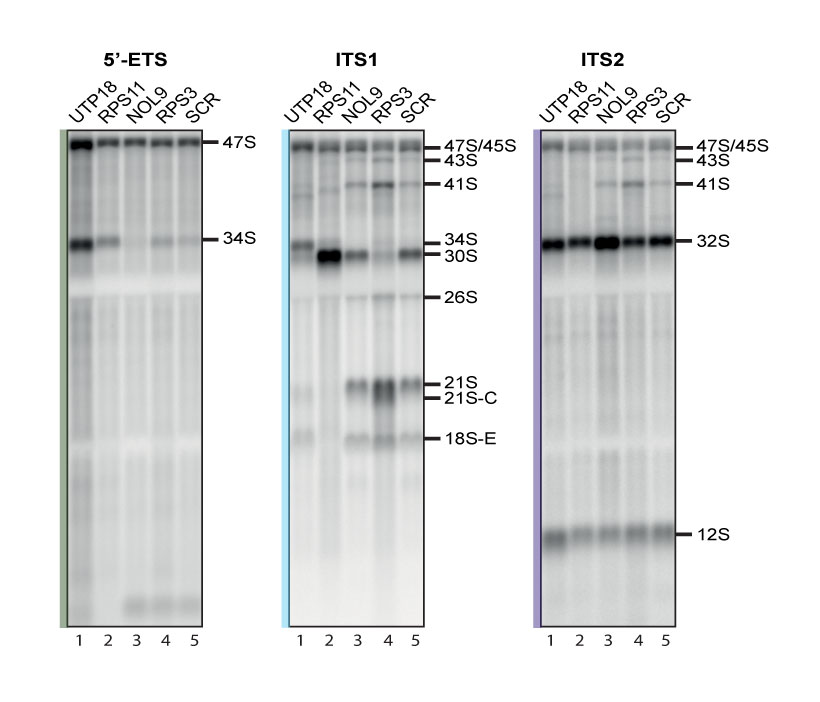
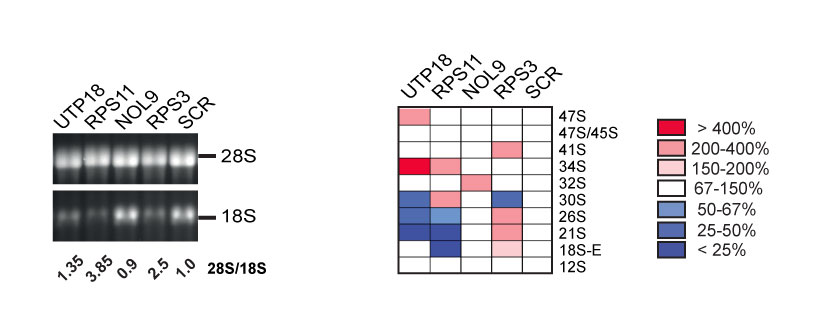
The calibration set is composed of 4 model genes: NOL9, RPS3, RPS11, and UTP18, as well as a non-targeting siRNA (SCR, for scrambled).
These genes were carefully selected for their known involvement in specific reactions in the pre-rRNA processing pathway [1,2,3]. For example, cells depleted for UTP18 accumulate the aberrant 34S RNA, resulting from inhibitions of early cleavage reactions (at sites 01, A0 and 1). Cells deprived of RPS11 strongly accumulate the 30S pre-rRNA, which is indicative of inhibition of cleavage at A0 and 1. NOL9 is primarily involved in ITS2 processing and in its absence the 32S pre-rRNA is accumulated. Finally, RPS3 was chosen mostly because it accumulates the 43S and the 26S pre-rRNAs, revealing a substantial level of uncoupling of cleavage at sites A0 and 1 (see [4] for further discussion). Note that in our high-resolution analysis, cells deprived of RPS3 were also shown to accumulate the 21S-C, a shorter version of 21S and late precursor intermediate in the 18S rRNA pathway (see [4,5]). To our knowledge, this had not been documented previously.
The calibration set, which was systematically included in each batch of siRNA-mediated depletions, and loaded on each gel, allowed us to technically validate the siRNA inactivation and to unambiguously identify pre-rRNA intermediates.
References
- Holzel M, Orban M, Hochstatter J, Rohrmoser M, Harasim T, Malamoussi A, Kremmer E, Langst G, Eick D (2010) Defects in 18 S or 28 S rRNA Processing Activate the p53 Pathway. J Biol Chem 285: 6364-6370.
- O’Donohue MF, Choesmel V, Faubladier M, Fichant G, Gleizes PE (2010) Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol 190: 853-866.
- Heindl K, Martinez J (2010) Nol9 is a novel polynucleotide 5′-kinase involved in ribosomal RNA processing. EMBO J 29: 4161-4171.
- Mullineux ST, Lafontaine DL (2012) Mapping the cleavage sites on mammalian pre-rRNAs: Where do we stand? Biochimie PMID: 22342225.
- Carron C, O’Donohue MF, Choesmel V, Faubladier M, Gleizes PE (2011) Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res 39: 280-291.